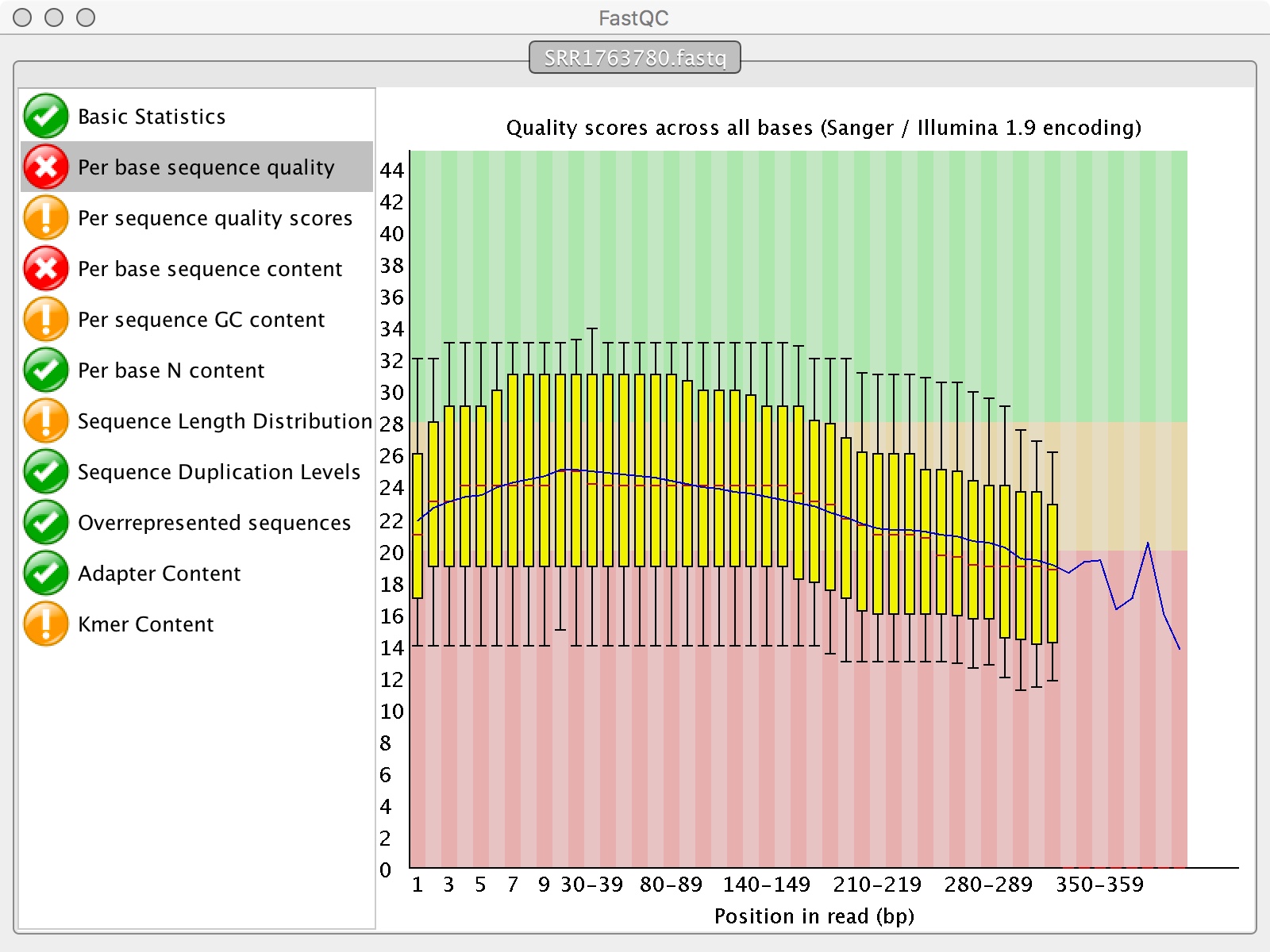
Lab 2: Genome Annotation and Comparison
Why use Prokka? Because in a benchmark test it has been shown to be as or more accurate at reproducing known annotation than RAST or xBASE2 in most annotation categories. It can run on a normal laptop for a small genome and is easy and convenient to use. Setup Prokka is not installed on the hpc-student cluster yet, so we’re going to learn how to install a program for your own use on the cluster. Jon Halter in University Research…